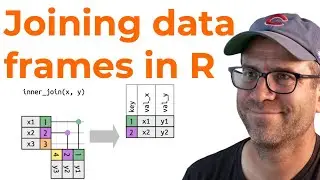
Finding the consensus classification using anonymous functions (CC284)
With 100 classification strings, we now need to find the consensus classification across those bootstrapped replicates. We'll make use of anonymous functions with the base R lapply and sapply functions. Of course we'll also make use of test driven development using the testthat package.This episode is part of an ongoing effort to develop an R package that implements the naive Bayesian classifier.
If you want to get a physical copy of R Packages: https://amzn.to/43pMR8L
If you want a free, online version of R packages: https://r-pkgs.org/
You can find my blog post for this episode at https://www.riffomonas.org/code_club/....
Check out the GitHub repository at the:
Beginning of the episode: https://github.com/riffomonas/phyloty...
End of the episode: https://github.com/riffomonas/phyloty...
#rstats #refactor #testthat #tdd #microbenchmark #vectors #rdp #16S #classification #classifier #microbialecology #microbiome
Support Riffomonas by becoming a Patreon member!
/ riffomonas
Want more practice on the concepts covered in Code Club? You can sign up for my weekly newsletter at https://shop.riffomonas.org/youtube to get practice problems, tips, and insights.
If you're interested in purchasing a video workshop be sure to check out https://riffomonas.org/workshops/
You can also find complete tutorials for learning R with the tidyverse using...
Microbial ecology data: https://www.riffomonas.org/minimalR/
General data: https://www.riffomonas.org/generalR/
0:00 Introduction
3:37 Game plan for classifying unknown sequences
10:26 Developing test for consensus function
14:28 Writing code to find consensus (Part 1)
16:58 Developing and testing consensus helper
23:07 Writing code to find consensus (Part 2)
Using real 16S rRNA gene sequence






![💖Korean Drama💖Kiss Goblin [키스요괴]💖Ban Sook X Oh Yeon Ah💖Love Story FMV](https://images.mixrolikus.cc/video/vOGrHlW1SiE)